About
At GCB Hub, we enhance the understanding of biomarkers and their causal connections with complex traits and diseases across global populations.
Science
Large-scale imputation models for multi-ancestry proteome-wide association analysis.
Proteome-wide association studies (PWAS) decode the intricate proteomic landscape of biological mechanisms for complex diseases. Traditional PWAS model training relies heavily on individual-level reference proteomes, restricting its capacity to harness the emerging summary-level protein quantitative trait loci (pQTL) data in the public domain. Here we introduced BLISS, a novel framework to train protein imputation models using only pQTL summary statistics. By leveraging extensive pQTL data from the UK Biobank, deCODE, and ARIC studies, we applied BLISS to develop large-scale European PWAS models covering 5,779 unique proteins. We further extended BLISS to integrate with small-scale non-European individual-level datasets, enabling the development of models tailored to Asian and African ancestries. We validated the performance of BLISS models through a systematic multi-ancestry analysis of over 2,500 phenotypes across five major genetic data resources. The newly identified protein-phenotype associations offer valuable insights into their cross-ancestry transferability, the contributions of different proteomic platforms, and the complementary perspectives provided by distinct genomic mapping approaches relevant to drug discovery.
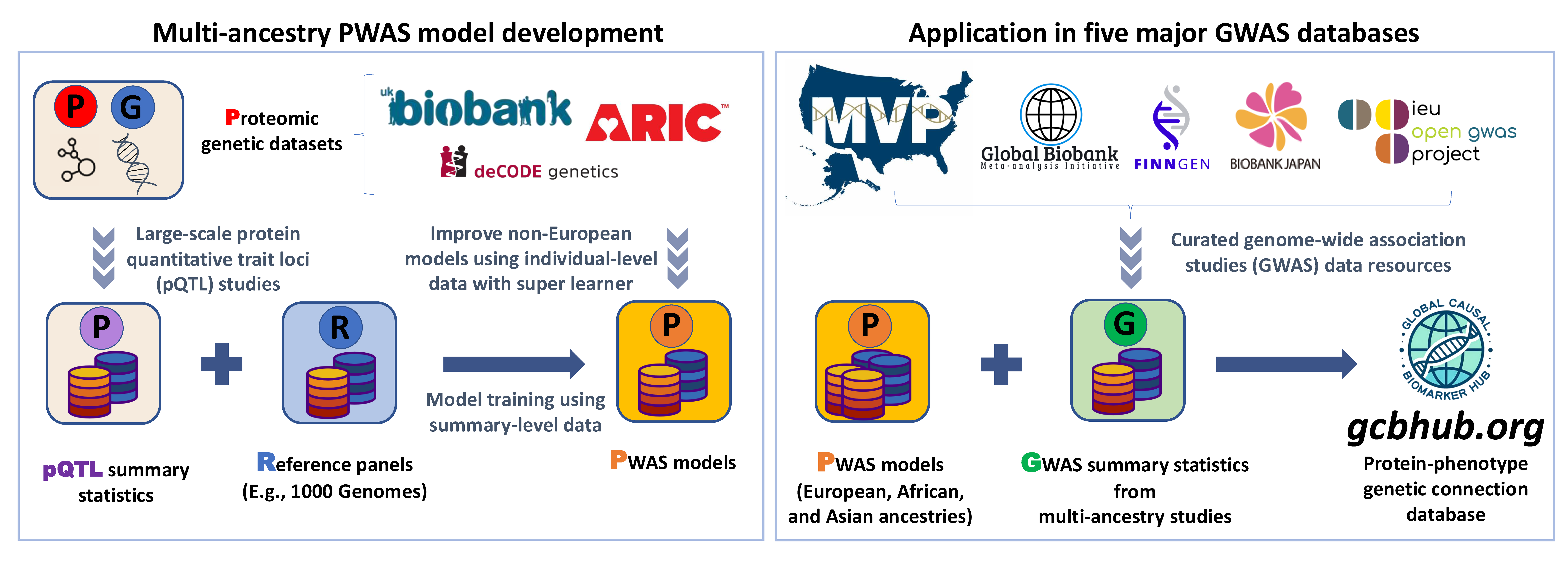
Note: BLISS code and developed models can be downloaded from our 🤗 Hugging Face repo. For the most current BLISS code, please refer to our GitHub page.
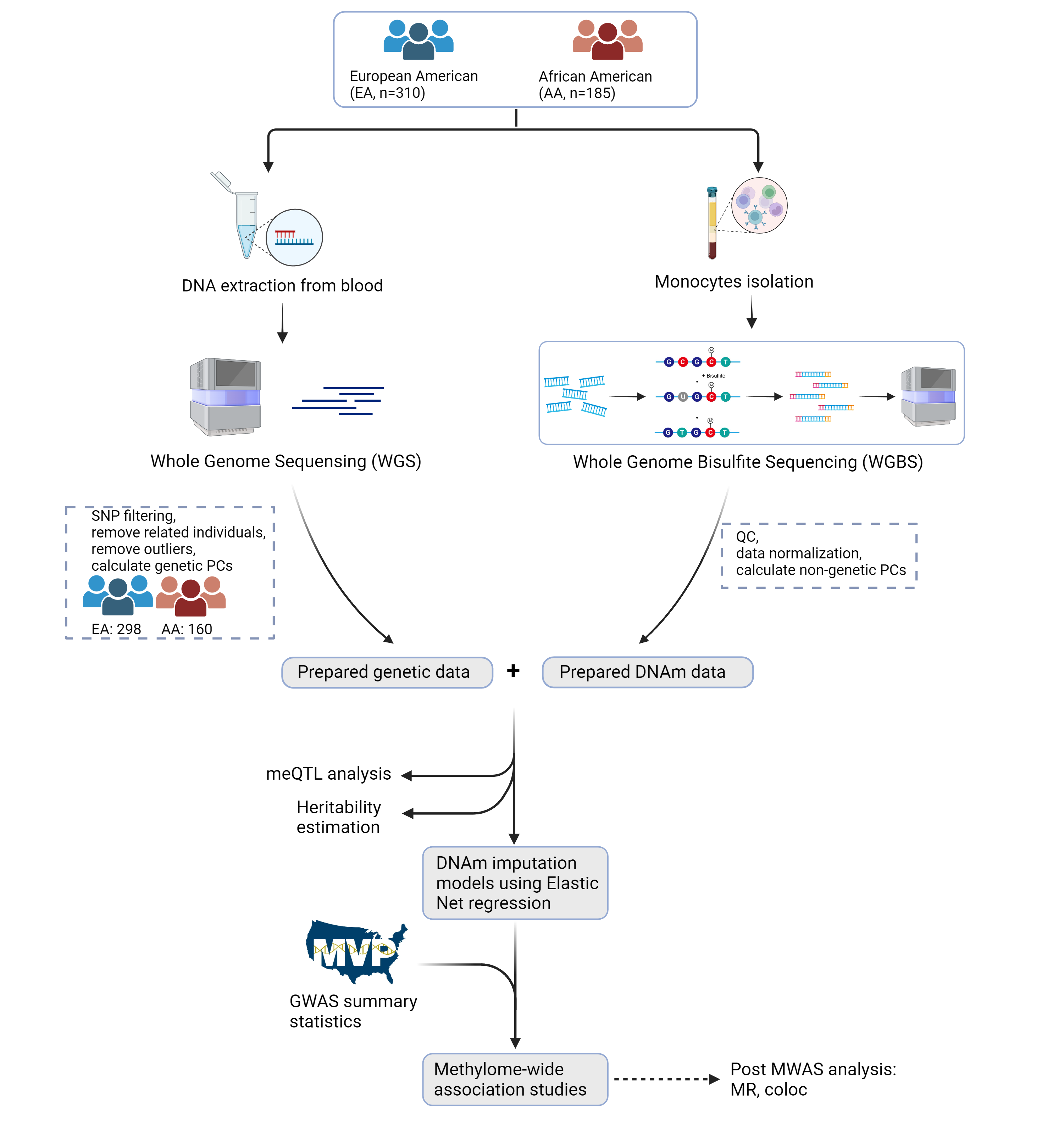
An atlas of genetic effects on the monocyte methylome across European and African populations.
Comprehensive Multi-Ancestry Methylome-Wide Association Study (MWAS)
This is a comprehensive multi-ancestry methylome-wide association study (MWAS) conducted on purified monocytes from European American (EA) and African American (AA) populations.
Key Features:
- Whole-genome bisulfite sequencing (WGBS) data from 298 EA and 160 AA individuals
- Analysis of over 25 million methylation sites
- Identification of cis- and trans-methylation quantitative trait loci (meQTLs)
- Development of population-specific DNA methylation imputation models
- MWAS analysis of 41 complex traits using Million Veteran Program (MVP) data
Our study provides:
- CpG-trait associations: Direct links between specific methylation sites and complex traits
- Gene-trait associations: Aggregated effects of methylation on genes associated with various phenotypes
This resource bridges the gap between genomics and the monocyte methylome, offering insights into:
- Genetic regulation of DNA methylation
- Novel methylation-phenotype associations
- Transferability of findings across diverse ancestries
The data presented here are valuable for researchers investigating the epigenetic basis of complex diseases, particularly those mediated by the immune system.
Team
GCB Hub aims to assemble a passionate team dedicated to open science, pulling together skills from a wide range of disciplines, including statistics, biostatistics, causal inference, epidemiology, drug development, clinical medicine, human genomics, as well as web and software development.
Leadership Team:
Chong Wu (MD Anderson) and Bingxin Zhao (UPenn)
Current and past members:
Zichen Zhang
![]() (MD Anderson),
Xiaochen Yang
(MD Anderson),
Xiaochen Yang
![]() (Purdue),
Wanheng Zhang
(Purdue),
Wanheng Zhang
![]() (MD Anderson)
(MD Anderson)
Contact and license
We are dedicated to identifying causal biomarkers, including but not limited to proteins, genes (expression and splicing), and CpG sites, for complex traits and diseases in different domains. Resulting resources would enable to address many relevant scientific questions, and help researchers in pinpointing the most promising targets for subsequent functional analysis, drug development, and repurposing.
If you have QTL datasets or would like to deposit your summary statistics to GCB Hub, please feel free to reach out to Chong Wu and Bingxin Zhao.
This work is licensed under the CC BY-NC-ND 4.0 DEED.